Abstract
Background: Human hematopoiesis was considered a hierarchical stepwise process, but on the basis of recent progress in single-cell RNA-sequencing (scRNA-seq) it is now considered a continuous process. However, the uncertainty or fluctuation of single-cell transcriptome dynamics during differentiation was not considered. To address this issue, we developed a new computational methodology that converts single-cell splicing kinetics into an advection-diffusion model in a latent cell state using deep learning technology, which enabled us to quantitatively capture the fluctuation in cell state dynamics. Because the human lymphoid pathway and bifurcation points for dendritic cells (DC)/monocytic lineage remain incompletely defined, we applied our method to explore the lymph-DC pathway in the human lympho-hematopoietic system.
Materials and methods: CD34+ or CD34+CD38+CD45RA+CD19-CD7-CD10+ cells (CD10SPs) were sorted from a mixture of frozen human cord blood samples, and scRNA-seq and single-cell chromatin accessibility analysis (scATAC) were performed. The data were applied to the variational inference of cell state dynamics with fluctuation (VICDYF). Subsequently, the obtained findings were validated by novel mesenchymal stem cell (MSC)-like coculture system that supports comprehensive human hematopoiesis.
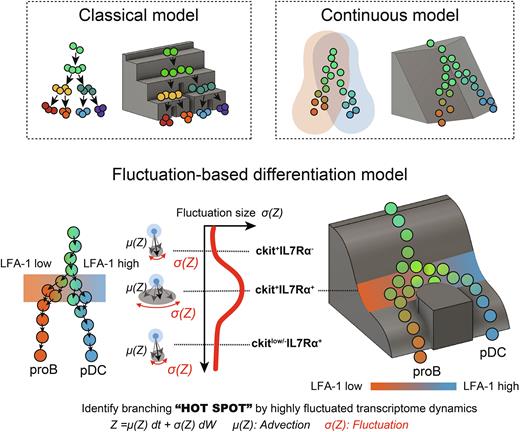
Results: MSC-like cultures of various lymphoid populations showed that the CD10SP population, which had been thought to be B/NK progenitors, was a heterogenous population in which lineage diversification from multi-lymphoid progenitors toward lymphoids and DCs was suspected to occur. Our insilico analysis of CD34+ cells with VICDYF suggested a B/plasmacytoid DC (pDC) bifurcation with high fluctuating transcriptome dynamics in the CD10-expressing fraction among various levels of fluctuating bifurcation points. Analysis of CD10SPs identified the B/pDC bifurcation as a highly fluctuating transcriptome state in the IL-7Rα+ fraction in association with the LFA-1 expression dynamics and pDC differentiation bias. Consistent with these findings, B/pDC common progenitors were identified by single cell culture, and the LFA-1-high population gave rise to a higher percentage of pDCs than proB cells, while the LFA-1-low population produced a higher percentage of proB cells than pDCs in the c-kit+IL-7Rα+ fraction. Of note, the epigenetic regulator SFMBT2 was specifically expressed in the B/pDC bifurcation region. The scATAC of the CD34+ population revealed that the B/pDC bifurcation with large fluctuation coincided with the high accessibility of both sides of the differentially accessible chromatin regions for both B and pDC lineages. This study revealed a new landscape of the human lymphoid pathway that includes crucial bifurcation points with highly fluctuating single-cell transcriptome dynamics.
Discussion: Our computational methodology discovered a novel B/pDC bifurcation as high fluctuating dynamics. The insilico predictions were confirmed by the biological findings, using MSC-like cultures. Although the underlying molecular mechanism remains elusive, single-cell multiomics analysis suggests that the estimated large fluctuation is associated with epigenetic changes in chromatin regions for both B and pDC lineages. VICDYF also identified the highly fluctuated transcriptome dynamics at the putative lineage bifurcation point during pancreatic development.
Conclusion: Our results provide a new model of differentiation - fluctuation based differentiation - which reconciles continuous and stepwise models.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal